RNA virus
https://en.wikipedia.org/wiki/RNA_virus
"An RNA virus is a virus which has (ribonucleic acid) RNA as its genetic material.[1] The nucleic acid is usually single-stranded RNA (ssRNA) but it may be double-stranded RNA (dsRNA).[2] Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, MERS, COVID-19, Dengue Virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio, mumps, and measles."
Messenger RNA genetics
BY The Editors of Encyclopaedia Britannica
https://www.britannica.com/science/messenger-RNA
"Messenger RNA (mRNA), molecule in cells that carries codes from the DNA in the nucleus to the sites of protein synthesis in the cytoplasm (the ribosomes). The molecule that would eventually become known as mRNA was first described in 1956 by scientists Elliot Volkin and Lazarus Astrachan. In addition to mRNA, there are two other major types of RNA: ribosomal RNA (rRNA) and transfer RNA (tRNA)."
"Because information in DNA cannot be decoded directly into proteins, it is first transcribed, or copied, into mRNA (see transcription). Each molecule of mRNA encodes the information for one protein (or more than one protein in bacteria), with each sequence of three nitrogen-containing bases in the mRNA specifying the incorporation of a particular amino acid within the protein. The mRNA molecules are transported through the nuclear envelope into the cytoplasm, where they are translated by the rRNA of ribosomes (see translation)."
RNA Functions
By: Suzanne Clancy, Ph.D. © 2008 Nature Education
Citation: Clancy, S. (2008) RNA Functions. Nature Education 1(1):102
"The central dogma of molecular biology suggests that DNA maintains the information to encode all of our proteins, and that three different types of RNA rather passively convert this code into polypeptides. Specifically, messenger RNA (mRNA) carries the protein blueprint from a cell's DNA to its ribosomes, which are the "machines" that drive protein synthesis. Transfer RNA (tRNA) then carries the appropriate amino acids into the ribosome for inclusion in the new protein. Meanwhile, the ribosomes themselves consist largely of ribosomal RNA (rRNA) molecules."
SPARS Pandemic 2025–2028
https://de.wikipedia.org/wiki/SPARS_Pandemic_2025%E2%80%932028
"SPARS Pandemic 2025–2028 ist die Bezeichnung für eine Pandemiesimulation, die im Oktober 2017 abgeschlossen wurde. Sie wurde vom Center for Health Security der Johns Hopkins University durchgeführt. Die Übung stellte in einem "futuristischen Szenario" dar, welche Kommunikationsdilemmata hinsichtlich medizinischer Maßnahmen in nicht allzu ferner Zukunft entstehen könnten.[1]
Die Übung war eines von vier Planspielen in den USA, die teilweise Ereignissen der späteren COVID-19-Pandemie entsprachen, die drei anderen waren Atlantic Storm, Clade X und Event 201."
FOR IMMEDIATE RELEASE
https://www.centerforhealthsecurity.org/event201/191017-press-release.html
"EVENT 201 PANDEMIC EXERCISE UNDERSCORES IMMEDIATE NEED FOR GLOBAL PUBLIC-PRIVATE COOPERATION TO MITIGATE SEVERE ECONOMIC AND SOCIETAL IMPACTS OF PANDEMICS
Johns Hopkins Center for Health Security, World Economic Forum and Bill & Melinda Gates Foundation host Event 201 in New York, and a Virtual Exercise
NEW YORK, Updated Oct. 17, 2019 – The Johns Hopkins Center for Health Security, with the World Economic Forum and the Bill & Melinda Gates Foundation, will host Event 201, a multimedia global pandemic exercise on Friday, Oct. 18, 2019, in New York City. The public may register and participate in the simultaneous virtual exercise in English, 8:50 a.m.-12:30 p.m. EDT at centerforhealthsecurity.org/event201/. The exercise underscores the need for global public-private cooperation to mitigate economic and societal impacts of severe pandemics.
In recent years, the world has seen a growing number of epidemic events, about 200 per year, which strain limited resources. A large global pandemic would be disruptive to health, economies, and society. Economic studies show that pandemics could cause an average annual economic loss of 0.7% of global GDP—or $570 billion.
“In addition to challenging health systems, pandemics can cause severe cascading economic and societal consequences,” said Tom Inglesby, MD, director of the Johns Hopkins Center for Health Security at the Bloomberg School of Public Health, “Neither governments nor private industries alone can adequately respond to a severe pandemic; they must work together. We’ve designed Event 201 to engage leaders in compelling ways to help them understand the decisions needed to prepare for and respond to biological threats.
Event 201, played by 15 leaders of business, government, and public health, will illustrate realistic policy problems that must be addressed under pressure during a pandemic. At the video-driven exercise, players will be presented with a scenario that reveals unresolved and controversial policy and economic issues that could be solved with sufficient political will, financial investment, and attention. Players include:
Latoya Abbott, Risk Management & Global Senior Director Occupational Health Services, Marriott International
Sofia Borges, Senior Vice President, UN Foundation
Brad Connett, President, U.S. Medical Group, Henry Schein, Inc.
Christopher Elias, President, Global Development division, Bill & Melinda Gates Foundation
Tim Evans, Former Senior Director of Health, World Bank Group
George Gao, Director-General, Chinese Center for Disease Control and Prevention
Avril Haines, Former Deputy Director, Central Intelligence Agency; Former Deputy National Security Advisor
Jane Halton, Board member, ANZ Bank; Former Secretary of Finance & Former Secretary of Health, Australia
Matthew Harrington, Global Chief Operations Officer, Edelman
Martin Knuchel, Head of Crisis, Emergency and Business Continuity Management, Lufthansa Group Airlines
Eduardo Martinez, President, The UPS Foundation
Stephen Redd, Deputy Director for Public Health Service and Implementation Science, US CDC
Hasti Taghi, Vice President & Executive Advisor, NBCUniversal Media
Adrian Thomas, Vice President, Global Public Health, Johnson & Johnson
Lavan Thiru, Chief Representative, Monetary Authority of Singapore
“Outbreaks of infectious disease are inevitable, but the economic damage they cause is not,” said Ryan Morhard, project lead for Global Health Security at the World Economic Forum. “Sustained attention from a broad multistakeholder coalition is needed in advance of a severe pandemic to save lives and minimize economic and societal consequences.”
Chris Elias, president of global development at the Gates Foundation, noted that “Event 201 and its predecessor simulations like Clade X are crucial tools to understand not only what is needed to effectively respond to global public health crises, but also the consequences of what happens when we are not prepared.”
The exercise is supported by funding from the Open Philanthropy Project.
More information is at centerforhealthsecurity.org/event201, #Event201, @JHSPH_CHS, @wef and @gatesfoundation.
EVENT 201 IS A FICTIONAL EXERCISE AND DISEASE
# # #
About the Johns Hopkins Center for Health Security:
The Johns Hopkins Center for Health Security works to protect people from epidemics and disasters and build resilient communities through innovative scholarship, engagement, and research that strengthens the organizations, systems, policies, and programs essential to preventing and responding to public health crises. The Center is part of the Johns Hopkins Bloomberg School of Public Health and is located in Baltimore, MD.
About the World Economic Forum:
As the International Organization for Public-Private Cooperation, the World Economic Forum is committed to managing risks associated with emerging infectious diseases of epidemic and pandemic potential through innovative, cross-industry, and cross-sectoral public-private cooperation, strengthening national and global health security. Via the Forum’s Epidemics Readiness Accelerator, more than 100 stakeholders are addressing challenges associated with public-private cooperation relied upon for effective readiness. The 2019 Global Risk Report describes the transformation of biological risks, and the 2019 report, “Outbreak Readiness and Business Impact,” helps companies properly understand risks, enabling them to reduce their exposure, improve their resilience, and deliver on key opportunities for public-private cooperation to strengthen global health security.
About the Bill & Melinda Gates Foundation:
Guided by the belief that every life has equal value, the Bill & Melinda Gates Foundation works to help all people lead healthy, productive lives. In developing countries, it focuses on improving people’s health and giving them the chance to lift themselves out of hunger and extreme poverty. In the United States, it seeks to ensure that all people—especially those with the fewest resources—have access to the opportunities they need to succeed in school and life. Based in Seattle, Washington, the foundation is led by CEO Sue Desmond-Hellmann and Co-chair William H. Gates Sr., under the direction of Bill and Melinda Gates and Warren Buffett.
Editor’s note: RSVP and receive information about camera and recording limitations. Video, graphics, audio, photos and interviews will be available immediately after the exercise. See Event 201 media advisory and other materials.
Media Contact:
Margaret Miller
Director of Communications
Johns Hopkins Center for Health Security
centerhealthsecurity@jhu.edu"
Genotypic Diversity and Epidemiology of Human Rhinovirus Among Children With Severe Acute Respiratory Tract Infection in Shanghai, 2013–2015
Front. Microbiol., 07 August 2018 | https://doi.org/10.3389/fmicb.2018.01836
1Key Laboratory of Laboratory Medicine, Ministry of Education, Institute of Medical Virology, Wenzhou Medical University, Wenzhou, China
2National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
3Infectious Disease Department, Children’s Hospital of Fudan University, Shanghai, China
Introduction
Human rhinovirus (HRV), a single-stranded positive-sense RNA virus, belongs to the genus Enterovirus and family Picornaviridae, and is classified as HRV-A, -B, or -C. HRV-A and HRV-B were discovered in the 1950s (Price, 1956), while HRV-C was identified using molecular techniques in 2006 (Lamson et al., 2006; Lau et al., 2007). According to the 2017 International Committee on the Taxonomy of Viruses (ICTV) release1, a total of 168 HRV genotypes (80 HRV-A, 32 HRV-B, and 56 HRV-C) are recognized (Kuroda et al., 2015).
Human rhinovirus genomic RNA is approximately 7.2 kb that consisting of a single open reading frame (ORF) encodes 11 proteins, with 5′ and 3′ untranslated regions (UTR) at both end, respectively. The ORF encodes a poly-protein which is cleaved by viral proteases to produce 11 proteins including four structural viral proteins (VP) 1 to 4. Compared to the rest of the HRV genome, the capsid proteins exhibit a high degree of heterogeneity resulting in a wide range of antigenic diversity. RT-PCR assays targeted the 5′-UTR are usually used for HRV clinical detection. HRV species and types are classified almost exclusively now based on VP1 or VP4/VP2 sequence alignments (Wisdom et al., 2009).
Human rhinovirus is a frequently detected respiratory virus in children with mild acute respiratory infection (ARI), but may also lead to more-severe respiratory tract symptoms, such as pneumonia, bronchiolitis, and asthma. HRV is, after respiratory syncytial virus (RSV), the second most frequent viral cause of community-acquired pneumonia and other severe acute respiratory infections (SARIs) (Honkinen et al., 2012; Esposito et al., 2013). HRV-C is more frequently associated with wheezing episodes, asthma exacerbations, and lower respiratory tract infections compared with HRV-A and -B (Linsuwanon et al., 2009; Gern, 2010; Bizzintino et al., 2011). However, there is reportedly no relationship between disease severity and HRV species (Lee et al., 2012; Chen et al., 2015; van der Linden et al., 2016).
Data on the genotypic diversity and epidemiology of HRVs in children with SARI are sparse. Thus, we evaluated the predominant HRV species and genotypes, and their associations with the clinical characteristics, of 1,003 children hospitalized with SARI from 2013 to 2015 in Shanghai, China.
Materials and Methods
Ethics Issues
All aspects of the study were performed in accordance with the national ethics regulations and approved by the Ethics Committee of the Children’s Hospital of Fudan University (Jun Shen; CHFU2013016) as well as the Ethics Committee of National Institute for Viral Disease Control and Prevention (RL; IVDC2013022). Participants were received “Written Informed Consent” on the study’s purpose and of their right to keep information confidential. Written consent was obtained from all participants or their guardians.
Patients and Sample Collection
From June 2013 to August 2015, 1,003 nasopharyngeal aspirates (NPAs) were collected from children hospitalized with SARI in the Children’s Hospital of Fudan University, Shanghai, China. The revised World Health Organization SARI case definition (World Health Organization [WHO], 2011) was adopted and cases with clinical suspicion of SARI for children was enrolled (Zhang et al., 2013; Wang et al., 2016). Eligibility and classification of the clinical syndromes of SARI were determined from individual’s original record of medical history and examination. The criteria of hospitalized patient inclusion were: sudden onset of fever >38°C and cough or sore throat and difficulty breathing (dyspnea, oxygen saturation < 90%). Additional criteria were a normal or low leukocyte count, or lower chest wall indrawing. Demographic data and clinical findings at the time of diagnosis were recorded on a standard form. All NPA samples were stored at -80°C until use.
HRV Detection and Genotyping
Viral nucleic acid was extracted from 200 μL of sample using QIAamp MinElute Virus Spin Kits (Qiagen, Germany). cDNA was synthesized using an AMV reverse transcriptase and random hexamer primers (Promega, United States), as described previously (Lu et al., 2012). Nested RT-PCR targeting of the 5′-UTR was employed for HRV screening, and of the VP4–VP2 regions for genotyping. All (totally nine, except HEV IS only) of HRV or HRV/HEV primers from 5-UTR to VP4–VP2 were used to detect HRV, as described previously (Wisdom et al., 2009). Specimens from which amplification of the VP4–VP2 regions failed were defined as untyped. PCR products were confirmed by sequencing. Phylogenetic analysis was conducted using Molecular Evolutionary Genetics Analysis (MEGA) software (ver. 7).
Detection of Common Respiratory Viruses
Human rhinovirus-positive specimens were screened for influenza virus types A and B, parainfluenza virus types 1 to 3, RSV, picornaviruses (enteroviruses and rhinoviruses), and adenovirus (AdV) using three multiple-nested-PCR assays; and for human metapneumovirus (hMPV), human bocavirus (HBoV), and human coronavirus 229E/OC43/NL63/HKU1 using nested-PCR assays. The multiple-nested and nested PCRs were performed as described previously (da Silva Filho et al., 2012; Lu et al., 2012). All PCR products of 15 common respiratory viruses were confirmed by sequencing.
Statistical Analysis
Data analysis was performed using PASW (ver. 18; SPSS Inc., United States) and GraphPad Prism software (ver. 5.0; GraphPad Software, Inc., United States). Age, maximum body temperature, laboratory parameters, clinical features, and HRV prevalence were compared by χ2-test or Fisher’s exact test for categorical variables, and by two-tailed paired Student’s t-test for continuous variables. A value of P < 0.05 was considered indicative of statistical significance.
Results
Epidemiology of HRV
In total, 1,003 NPAs were collected. The male: female ratio was 609:394 (1.55:1) and the median age was 1 year (range: 10 days to 15 years). HRV was detected in 280 (27.9%) of the 1,003 specimens: HRV-A in 140 (14.0%), HRV-B in 21 (2.1%), HRV-C in 56 (5.6%), and HRV-untyped in 63 (6.3%). The HRV detection rates are shown in Table 1. The detection rates of HRV-A and -B differed significantly (P < 0.001). In general, the 5′-UTR region highly conserved between HRV and enterovirus, causing cross-reactivity in RT-PCR assay for the two viruses. In the current study, 42 enterovirus positive samples also were detected by nested RT-PCR targeting of the HRV 5′-UTR.
TABLE 1

TABLE 1. Population demographic of HRV-positive specimens.
Human rhinovirus infected significantly more males than females (65.7% vs. 34.3%, P = 0.044), and HRV was detected in patients of all ages, although HRV-B was not detected in those aged 3–4 years (Table 1). The detection frequency of HRV-A was significantly lower in patients aged 5–9 years (P = 0.008), but the frequencies of detection of HRV-B, -C, and -untyped did not differ according to age.
The seasonal distribution of HRV is shown in Figure 1. HRV-A was detected most frequently and peaked in winter 2013 and autumn 2015. HRV-C detection peaked in autumn (17.1% in 2013 and 20.7% in 2014). HRV-B was detected year-round at similar frequencies in all seasons. Detection of HRV-untyped peaked in summer and spring 2014 and was at a low level thereafter.
FIGURE 1
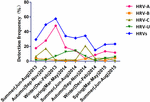
FIGURE 1. Temporal distribution of the HRV species between June 2013 and August 2015 inclusive.
Genotypic Diversity of HRV
Sequence analysis based on 420 bp of the VP4–VP2 region of 217 of 280 the HRV strains yielded 77 genotypes: 43 HRV-A (most frequently detected: -A78 [17/140, 12.1%], followed by -A12 [15/140, 10.7%], -A89 [8/140, 5.7%], -A61 [N = 6], and -A1, -A22, -A56, and A-58 [N = 5 each]), 10 HRV-B (most frequently detected: -B70 [5/21, 23.8%], followed by -B86 and -B97 [3/21, 14.3%]) and 24 HRV-C (most frequently detected: -C2, -C6, and -C24, [6/56, 10.7%], followed by -C16 [5/56, 8.9%] and -C4 and -C53 [4/56, 7.1%]) (Figure 2).
FIGURE 2
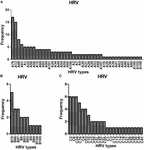
FIGURE 2. Detection of HRV genotypes in the study (A–C).
Clinical Characteristics
The clinical characteristics of the patients with HRV infections are listed in Table 2. All HRV-positive patients presented with SARI. Fever, cough, gasping, cyanosis, and diarrhea were the most common symptoms at presentation. There was no significant difference in clinical symptoms according to HRV species, except that cough and cyanosis were less frequent in patients infected with HRV-A and HRV-C, respectively (P = 0.048 and 0.005).
TABLE 2

TABLE 2. Clinical characteristics of patients with HRV infections in the study.
Co-infection With Other Respiratory Viruses
Of the 280 HRV-positive patients, 131 (46.8%) were co-infected—102 with one other virus, 21 with two other viruses, and 8 with three other viruses (Table 3). Most such co-infections involved AdV, HBoV, and RSV (39.7, 38.2, and 17.6%, respectively). The patients with co-infections did not have more serious disease (data not shown).
TABLE 3

TABLE 3. Co-detections of HRV and other respiratory virus in the study.
Discussion
We evaluated the HRV genotype distribution in children with SARI in the Children’s Hospital of Fudan University, Shanghai, China from June 2013 to August 2015. Of the 1,003 samples, 280 (27.9%) were positive for HRV (HRV-A, 50.0%; HRV-B, 7.5%; HRV-C, 20%; and HRV-untyped, 22.5%). This HRV detection rate is consistent with prior studies (11.0–40.6%) worldwide, as are the proportions of the three HRV species (HRV-A, 44.4–56%; HRV-B, 2–12%; and HRV-C, 25–45.3%) (Xiang et al., 2010; Henquell et al., 2012; Rahamat-Langendoen et al., 2013; Marcone et al., 2014; Tsatsral et al., 2015; Xiao et al., 2015; Milanoi et al., 2016; van der Linden et al., 2016; Blaschke et al., 2018). This prevalence of HRV-untyped is higher than in a recent study in Chongqing (13%) (Xiao et al., 2015), perhaps because our study involved patients with SARI. We achieved identical results using another nested-PCR detection primer also targeting the VP4–VP2 region (data not shown). Further research on whether these HRV-untyped contain new genotypes is warranted.
We detected 77 HRV genotypes in Shanghai from 2013 to 2015 (43 HRV-A, 10 HRV-B, and 24 HRV-C). The predominant HRV-A genotypes were A-78, A-12, A-89, and A-61; those of HRV-B were B-70, B-86, and B-97; and the predominant HRV-C genotypes were C-2, C-6, C-24, and C-16. Variation in the prevalent HRV genotype has been reported by others. In Amsterdam, 129 HRV genotypes were detected in inpatients and outpatients: A-12, A-78, and C-2 predominated (van der Linden et al., 2016). In Buenos Aires, 30 HRV genotypes were detected in children with acute respiratory infections: A-101, A-49, and C-10 predominated (Marcone et al., 2014). In Asia, 59, 36, and 28 HRV genotypes were detected in Mongolia, Beijing, and Cambodia, respectively: A-46, A-12, A-78, B-79, B-86, C-2, and C-36; A-12 and B42; and A-89, A-78, B-79, and C-6, respectively, predominated (Xiang et al., 2010; Naughtin et al., 2015; Tsatsral et al., 2015). Therefore, a large number of HRV genotypes circulate simultaneously, and some genotypes (such as A-12, A-78, and C-2) are the more prevalent types across worldwide although various predominant genotype patterns were reported geographically.
Human rhinovirus circulated throughout this 2-year study with peaks in winter 2013 and autumn 2014; HRV-C predominated in autumn. This seasonal pattern is similar to those reported previously (Xiang et al., 2010; Marcone et al., 2014; van der Linden et al., 2016). Also, HRV-A was detected most frequently in those <5 years old, while the frequency of detection of HRV-B and -C did not differ by age. In contrast, in Mongolia, HRV-A and -C are detected more frequently in younger children, and -B more frequently in older children (Tsatsral et al., 2015).
Human rhinovirus-A and HRV-C are reportedly associated with more severe illness (Miller et al., 2011; Fawkner-Corbett et al., 2016); however, others have reported no link between species and disease severity (Xiang et al., 2010; Henquell et al., 2012; Rahamat-Langendoen et al., 2013; Marcone et al., 2014). In this study, there were no significant differences in clinical symptoms among the three HRV species, except that cough and cyanosis were less frequent in patients with HRV-A and HRV-C, respectively. HRV-C infection in boys under 5 years old with acute asthma significantly increases the risk of moderate/severe exacerbations (Lambert et al., 2017). Moreover, of 570 HRV-positive hospitalized patients with community-acquired pneumonia, 57 (10%) had viremia; the vast majority (98.2%) of viremic patients were infected with HRV-C. The frequency of HRV viremia was higher in patients 1–2 years of age, and patients with viremia were more likely to have severe clinical symptoms, such as chest retraction, wheezing, and asthma (Lu et al., 2017). Therefore, particular HRV species may be associated with disease severity at specific situations which further research is warranted.
Almost half (46.8%) of the HRV-positive patients were co-infected with other respiratory viruses, most frequently ADV, HBoV, and hRSV. This may reflect the viruses in circulation at the time, consistent with previous reports (Xiang et al., 2010; van der Linden et al., 2016; Blaschke et al., 2018). Additionally, for those patients infected with 2–4 respiratory viruses, further research is necessary to demonstrate the dominant virus by qPCR positive with CT value and impact on disease severity.
In summary, we report for in detail the first time the variety of HRV genotypes in circulation and their distribution according to season and age group, as well as their clinical symptoms, in children hospitalized with SARI in China during a 2-year period. Our data shown that HRV was detected in 280 (27.9%) of the 1,003 NPAs from children with SARI in Shanghai, in which HRV-A and -C were predominate. Among the 77 HRV genotypes detected, A78, A12, A89, B70, C2, C6, and C24 were the most frequent. The HRV species responsible for SARIs differs according to season and age.
Author Contributions
RL, JS, and WT conceived and designed the experiments. YZ, RL, JS, BW, and GL performed the experiments. RL, JS, and WT analyzed the data. RL and WT wrote the manuscript.
Funding
This work was supported by the Control and Prevention of Major Infectious Disease of China (2017ZX10104001-002-003 and 2014ZX10004001-002), and the National Key Research and Development Program of China (2016YFD0500301 and 2016YFC1200200). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest."



No comments:
Post a Comment